On January 23, 2020, Hans Clevers from the Hubrecht Institute in the Netherlands published an article in the journal Cell, the so called "Snake Venom Gland Organoids"is a successful establishment of long-term organoid culture systems for the venom glands of several species of snakes. Reassembly of the transcriptome of Cape coral snakes revealed high levels of toxin transcripts in the organoids. Single-cell RNA sequencing of organoids and primary tissue identified distinct venom-expressing cells as well as proliferative cells expressing known mammalian stem cell markers, and the cellular composition of organoids and primary tissue was similar. Organoid cultures maintained regional heterogeneity in individual venom component expression, and the venom peptides were similar to those found in crude venom and possessed biological activity.
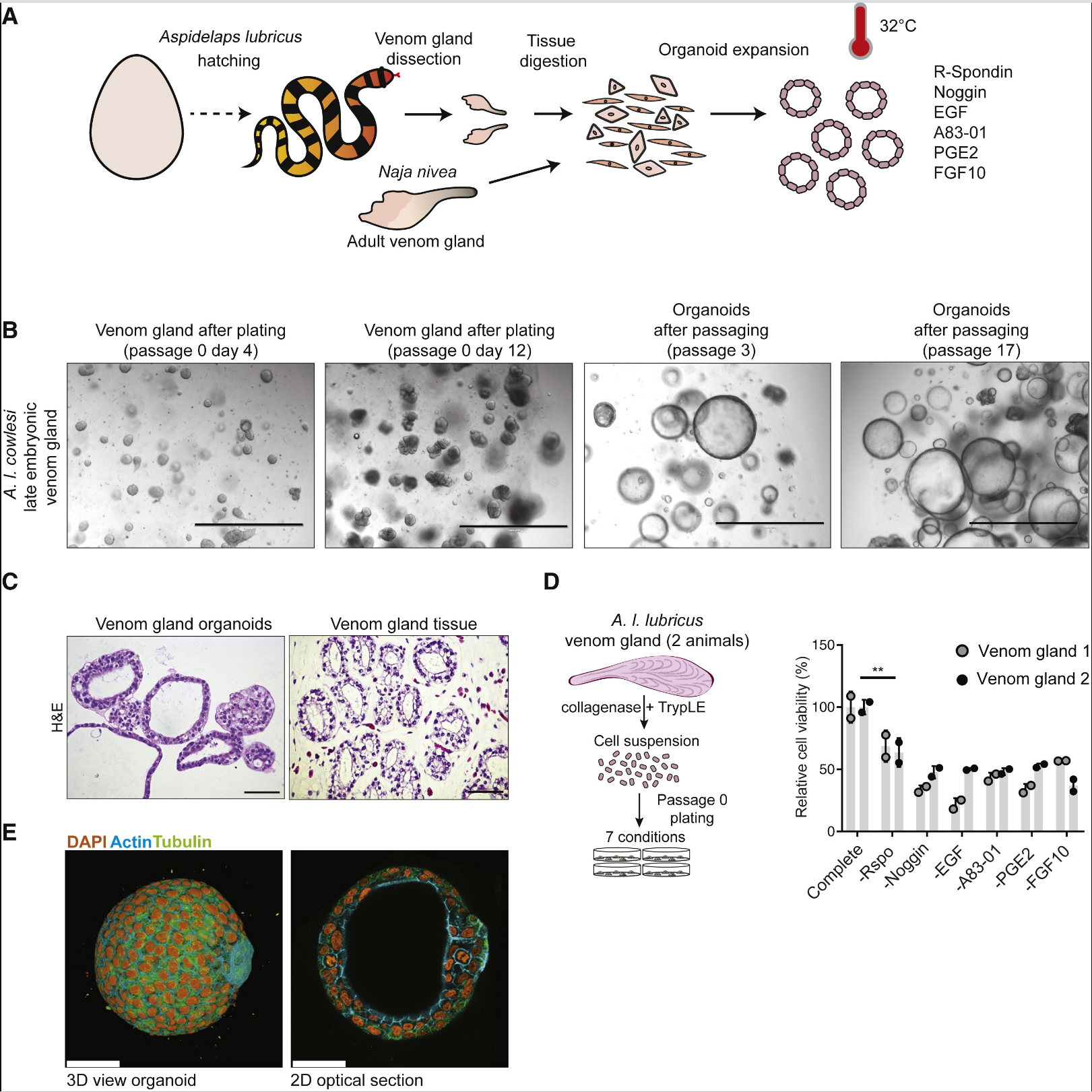
Researchers first isolated venom glands from nine different species of snakes at 32°C and embedded them in a basement membrane extract. Adding a “generic” mammalian organoid mixed medium led to the initial expansion of organoids from all snake venom glands. The expanded organoids produced by passaging resembled the original glandular epithelium histologically. Culturing with R-spondin (a Wnt signaling amplifier ligand for Lgr5), the BMP inhibitor Noggin, EGF, the small molecule TGF &beta inhibitor A83-01, PGE2, and FGF10 resulted in optimal amplification of the snake venom gland organoids, similar to the cell signaling pathways required for controlling mammalian epithelial organoids. Organoid culture required lower temperatures, consistent with the lower body temperature of snakes. Proliferating cells spontaneously organized into cystic spheroids with a simple polarized epithelium, and budding of organoids was occasionally observed. After more than 20 passages, there were no significant changes in growth kinetics or morphology of the organoids. Removing all growth factors led to reduced proliferation after seven days, and the organoids contained highly polarized secretory vesicles, showing a differentiated phenotype. The transcriptomes of the liver, pancreas, and organoids were compared with the reassembled reference transcriptome of A.I. cowlesi (a snake species), revealing that toxin expression was most prominent in the organoids, whereas the liver and pancreas corresponded to organ-specific markers. Among the toxins expressed by the organoids, 3FTxs were the most abundant, with CRISPs, SVMPs, and KUNs also being expressed, and the relative abundances of these toxins matched the transcriptional levels in the venom gland tissue and the protein levels in the crude venom. That is, A.I. cowlesi organoids expressed a spectrum of venom factors close to normal. To investigate the long-term passaging capability of adult venom gland organoids, researchers cultured organoids from the venom glands of adult Cape cobras (>1 year old), which not only reproduced the epithelial phenotype but could also be passaged for more than 18 generations, maintaining normal differentiation capacity and toxin expression.Single-cell RNA sequencing was performed on the expanded and differentiated organoids of A.I. cowlesi, as well as on primary tissues from late-stage embryonic venom glands. Twelve cell clusters were identified in the organoid cells, with the expanded and differentiated organoid cells largely separated. At least four cell clusters were identified as producing venom factors based on 3FTx expression, and venom-producing cells were predominantly found in the differentiated organoids. In the primary venom gland tissue cells, 20 cell clusters were identified, with gene expression indicating 53% epithelial cells, 27% stromal cells, 8% hematopoietic cells, 7% smooth muscle cells, and 4% endothelial cells. 3FTxs were most highly expressed in two epithelial cell clusters. Co-expression of PDI and 3FTxs was observed in both organoids and primary tissues. In the expanded organoids, toxin-expressing cells were mostly concentrated in cell clusters 6-10, with cell clusters 7, 8, and 9 expressing proliferation markers Ki-67 and mammalian stem cell markers RNF43, ASCL2, and LGR5, which were less common in the primary tissues. Addition of R-spondin3 increased the expression of stem cell markers and Wnt target genes, while addition of exogenous Wnt3a further increased the expression of these marker genes. Pseudotime analysis showed that cell clusters 7, 8, and 9, characterized by proliferative progenitor properties, were at the beginning of the pseudotime axis, while cell cluster 5 and cell clusters 1, 2, 3, and 4 were at the branches of differentiation.
Secretory cells in mammalian organs, such as the intestine, show regional isolation characteristics among different types of secretory cells. For A.I. cowlesi, when culturing region-specific organoids of the snake venom gland, it was found that the gene CTL was enriched in the “proximal” organoids, while “distal” organoids mainly expressed 3FTX and KUN toxins. The expression patterns were validated in primary venom gland tissues using RNA in situ hybridization. CRISP was uniformly expressed in both proximal and distal venom glands but was preferentially localized to basal rather than luminal cells. These data demonstrate regional heterogeneity in toxin expression within the venom gland. Seven days after differentiation, the presence of secretory vesicles and a marked accumulation of proteins in the lumen were detected, indicating that differentiated organoids produce large amounts of secretory proteins. Using LC-MS to compare extracts from organoids and crude venom, three major peaks with nearly identical masses between 6-8 kDa were found in both samples, likely corresponding to 3FTx and CRISP. The venom secreted by the organoids inhibited calcium signaling in cells, demonstrating its biological activity.
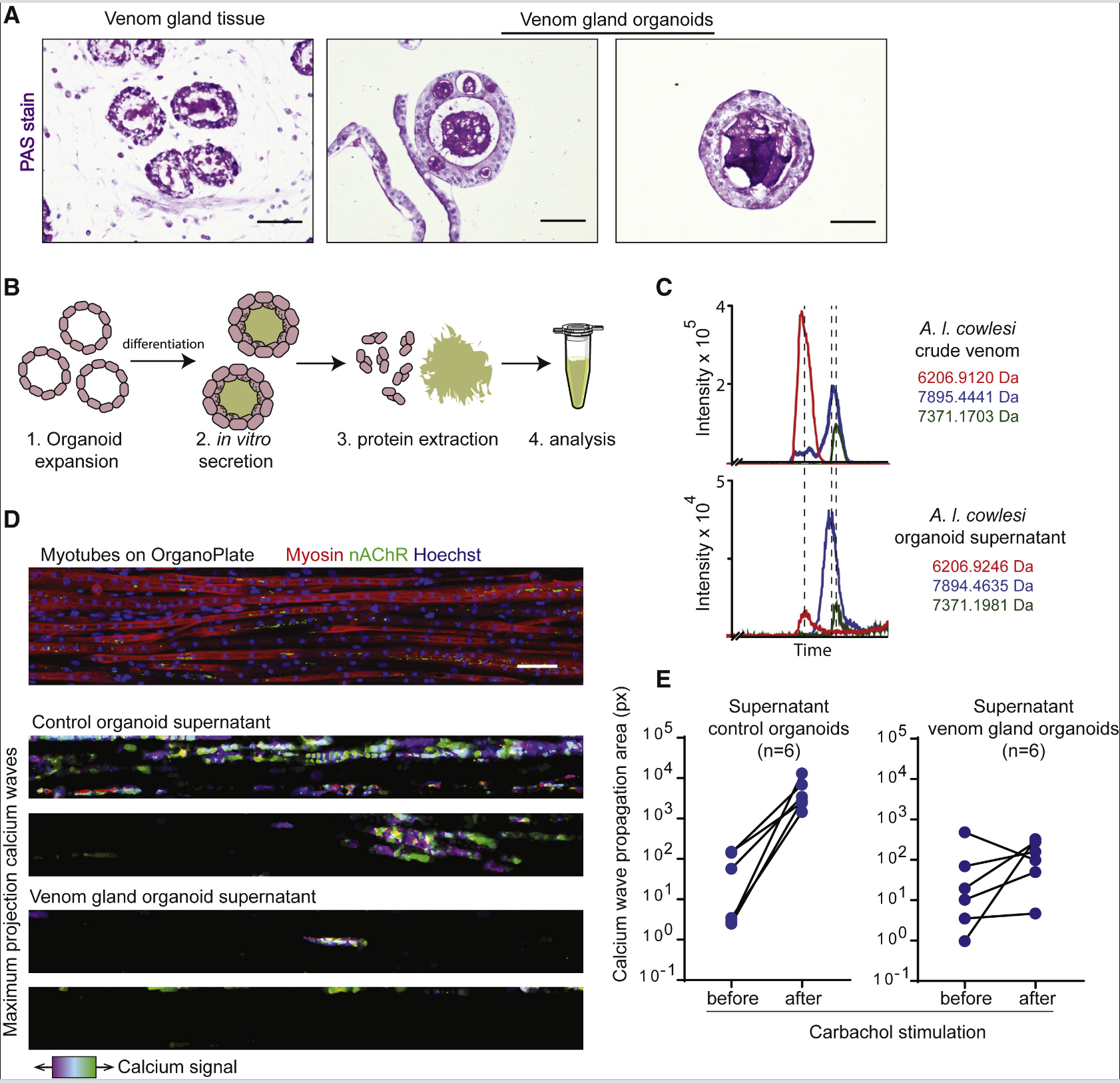
To sum up,The study extends organoid technology to reptilian tissues by establishing a long-term organoid culture system for snake venom glands. It demonstrates that the organoids possess stem cell properties, the ability for long-term passaging, and differentiation capabilities, essentially recapitulating the venom expression characteristics of the snake venom gland. The secreted venom shows biological activity, providing a new tool for studying venom glands.
1. Fry, B.G., Vidal, N., van der Weerd, L., Kochva, E., and Renjifo, C. (2009). Evolution and diversification of the Toxicofera reptile venom system. J. Proteomics 72, 127–136.
2. Sato, T., Vries, R.G., Snippert, H.J., van de Wetering, M., Barker, N., Stange, D.E., van Es, J.H., Abo, A., Kujala, P., Peters, P.J., and Clevers, H. (2009). Sin- gle Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265.
3. Maimets, M., Rocchi, C., Bron, R., Pringle, S., Kuipers, J., Giepmans, B.N., Vries, R.G., Clevers, H., de Haan, G., van Os, R., and Coppes, R.P. (2016). Long-Term In Vitro Expansion of Salivary Gland Stem Cells Driven by Wnt Signals. Stem Cell Reports 6, 150–162.





